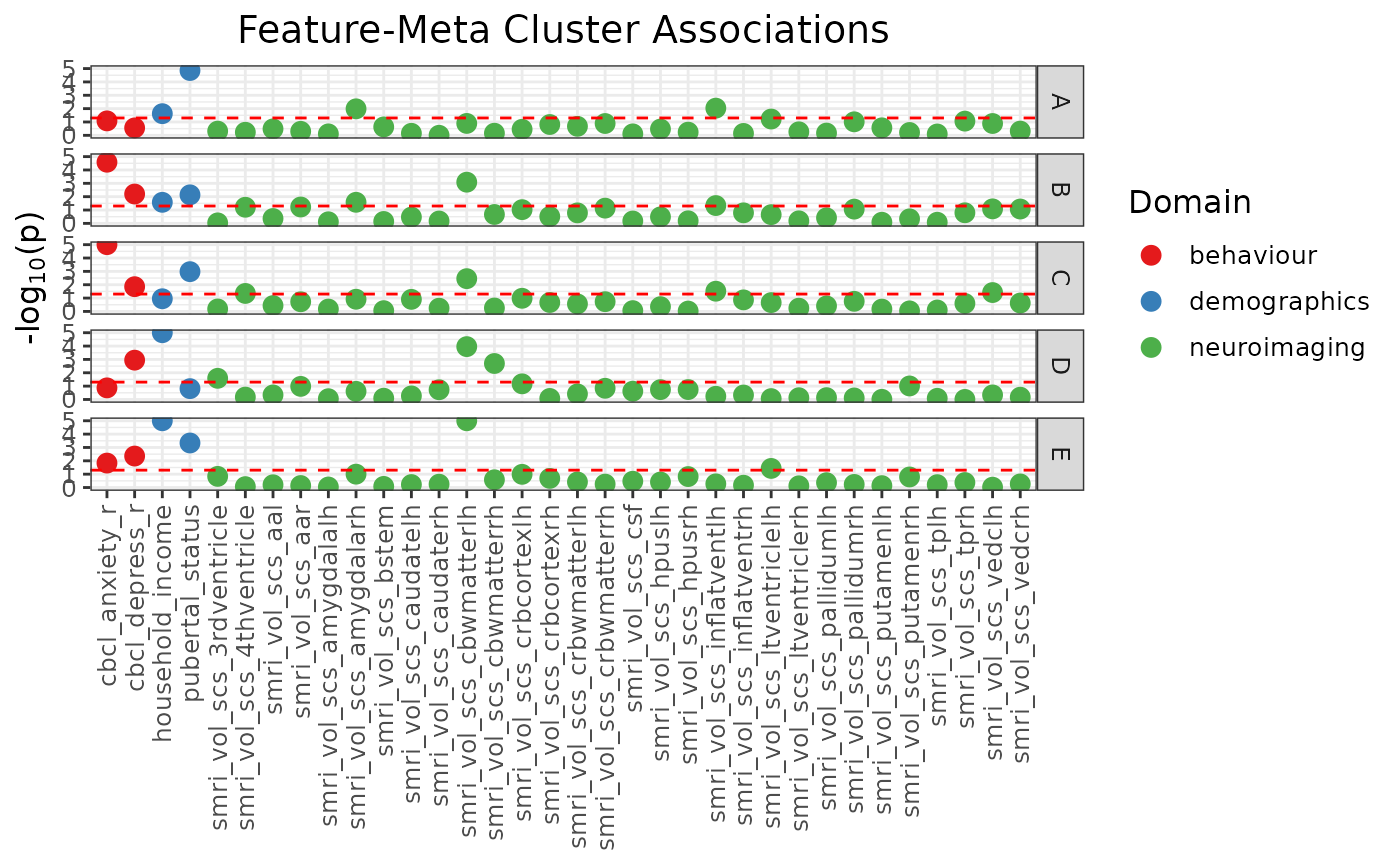
Manhattan plot of feature-meta cluster association p-values
Source:R/manhattan_plot.R
mc_manhattan_plot.RdGiven a data frame of representative meta cluster solutions (see
get_representative_solutions(), returns a Manhattan plot for showing
feature separation across all features in provided data/target lists.
Usage
mc_manhattan_plot(
ext_sol_df,
dl = NULL,
target_dl = NULL,
variable_order = NULL,
neg_log_pval_thresh = 5,
threshold = NULL,
point_size = 5,
text_size = 20,
plot_title = NULL,
xints = NULL,
hide_x_labels = FALSE,
domain_colours = NULL
)Arguments
- ext_sol_df
A sol_df that contains "_pval" columns containing the values to be plotted. This object is the output of
extend_solutions().- dl
List of data frames containing data information.
- target_dl
List of data frames containing target information.
- variable_order
Order of features to be displayed in the plot.
- neg_log_pval_thresh
Threshold for negative log p-values.
- threshold
p-value threshold to plot horizontal dashed line at.
- point_size
Size of points in the plot.
- text_size
Size of text in the plot.
- plot_title
Title of the plot.
- xints
Either "outcomes" or a vector of numeric values to plot vertical lines at.
- hide_x_labels
If TRUE, hides x-axis labels.
- domain_colours
Named vector of colours for domains.
Value
A Manhattan plot (class "gg", "ggplot") showing the association p-values of features against each solution in the provided solutions data frame, stratified by meta cluster label.
Examples
# \donttest{
dl <- data_list(
list(subc_v, "subcortical_volume", "neuroimaging", "continuous"),
list(income, "household_income", "demographics", "continuous"),
list(pubertal, "pubertal_status", "demographics", "continuous"),
list(anxiety, "anxiety", "behaviour", "ordinal"),
list(depress, "depressed", "behaviour", "ordinal"),
uid = "unique_id"
)
#> ℹ 188 observations dropped due to incomplete data.
sc <- snf_config(
dl = dl,
n_solutions = 20,
min_k = 20,
max_k = 50
)
#> ℹ No distance functions specified. Using defaults.
#> ℹ No clustering functions specified. Using defaults.
sol_df <- batch_snf(dl, sc)
ext_sol_df <- extend_solutions(
sol_df,
dl = dl,
min_pval = 1e-10 # p-values below 1e-10 will be thresholded to 1e-10
)
# Calculate pairwise similarities between cluster solutions
sol_aris <- calc_aris(sol_df)
# Extract hierarchical clustering order of the cluster solutions
meta_cluster_order <- get_matrix_order(sol_aris)
# Identify meta cluster boundaries with shiny app or trial and error
# ari_hm <- meta_cluster_heatmap(sol_aris, order = meta_cluster_order)
# shiny_annotator(ari_hm)
# Result of meta cluster examination
split_vec <- c(2, 5, 12, 17)
ext_sol_df <- label_meta_clusters(ext_sol_df, split_vec, meta_cluster_order)
# Extracting representative solutions from each defined meta cluster
rep_solutions <- get_representative_solutions(sol_aris, ext_sol_df)
mc_manhattan <- mc_manhattan_plot(
rep_solutions,
dl = dl,
point_size = 3,
text_size = 12,
plot_title = "Feature-Meta Cluster Associations",
threshold = 0.05,
neg_log_pval_thresh = 5
)
mc_manhattan
 # }
# }