Download a copy of the vignette to follow along here: correlation_plots.Rmd
In this vignette, we go through how you can visualize associations between the features included in your analyses.
Data set-up
library(metasnf)
# We'll just use the first few columns for this demo
cort_sa_minimal <- cort_sa[, 1:5]
# And one more mock categorical feature for demonstration purposes
city <- fav_colour
city$"city" <- sample(
c("toronto", "montreal", "vancouver"),
size = nrow(city),
replace = TRUE
)
city <- city |> dplyr::select(-"colour")
# Make sure to throw in all the data you're interested in visualizing for this
# data_list, including out-of-model measures and confounding features.
dl <- data_list(
list(cort_sa_minimal, "cortical_sa", "neuroimaging", "continuous"),
list(income, "household_income", "demographics", "ordinal"),
list(pubertal, "pubertal_status", "demographics", "continuous"),
list(fav_colour, "favourite_colour", "demographics", "categorical"),
list(city, "city", "demographics", "categorical"),
list(anxiety, "anxiety", "behaviour", "ordinal"),
list(depress, "depressed", "behaviour", "ordinal"),
uid = "unique_id"
)## ℹ 182 observations dropped due to incomplete data.
summary(dl)## name type domain length width
## 1 cortical_sa continuous neuroimaging 93 4
## 2 household_income ordinal demographics 93 1
## 3 pubertal_status continuous demographics 93 1
## 4 favourite_colour categorical demographics 93 1
## 5 city categorical demographics 93 1
## 6 anxiety ordinal behaviour 93 1
## 7 depressed ordinal behaviour 93 1
# This matrix contains all the pairwise association p-values
assoc_pval_matrix <- calc_assoc_pval_matrix(dl)
assoc_pval_matrix[1:3, 1:3]## mrisdp_303 mrisdp_304 mrisdp_305
## mrisdp_303 0.0000000 0.6374024 0.4513919
## mrisdp_304 0.6374024 0.0000000 0.2790341
## mrisdp_305 0.4513919 0.2790341 0.0000000Heatmaps
Here’s what a basic heatmap looks like:
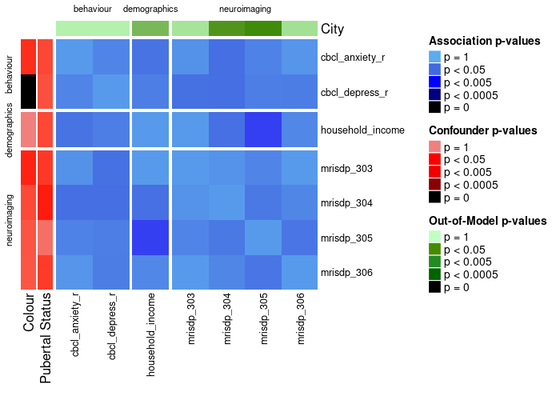
ap_heatmap <- assoc_pval_heatmap(assoc_pval_matrix)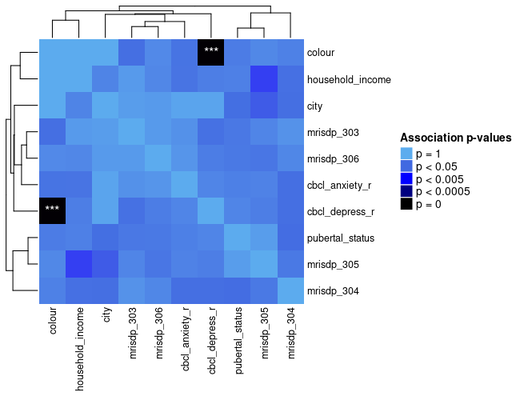
Most of this data was generated randomly, but the “colour” feature is really just a categorical mapping of “cbcl_depress_r”.
You can draw attention to confounding features and/or any out of model measures by specifying their names as shown below.
ap_heatmap2 <- assoc_pval_heatmap(
assoc_pval_matrix,
confounders = list(
"Colour" = "colour",
"Pubertal Status" = "pubertal_status"
),
out_of_models = list(
"City" = "city"
)
)
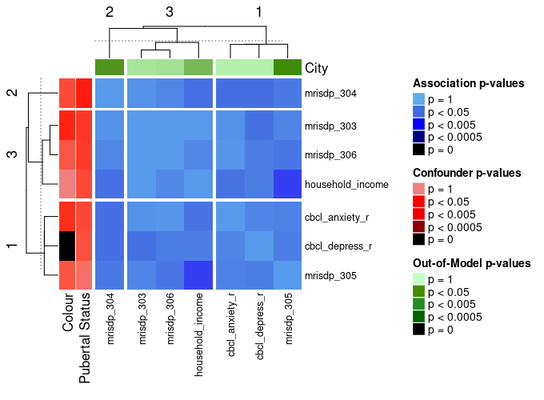
The ComplexHeatmap package offers functionality for splitting heatmaps into slices. One way to do the slices is by clustering the heatmap with k-means:
ap_heatmap3 <- assoc_pval_heatmap(
assoc_pval_matrix,
confounders = list(
"Colour" = "colour",
"Pubertal Status" = "pubertal_status"
),
out_of_models = list(
"City" = "city"
),
row_km = 3,
column_km = 3
)
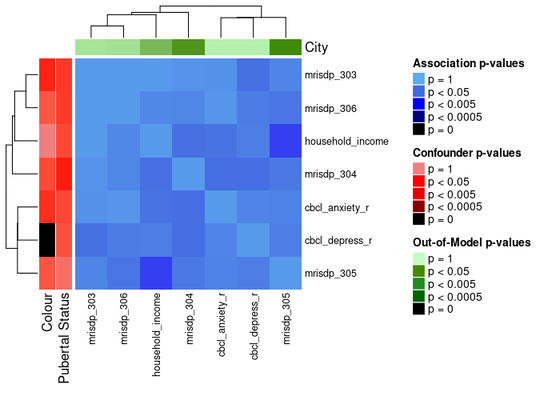
Another way to divide the heatmap is by feature domain. This can be
done by providing a data_list with all the features in the
assoc_pval_matrix and setting split_by_domain
to TRUE.
ap_heatmap4 <- assoc_pval_heatmap(
assoc_pval_matrix,
confounders = list(
"Colour" = "colour",
"Pubertal Status" = "pubertal_status"
),
out_of_models = list(
"City" = "city"
),
dl = data_list,
split_by_domain = TRUE
)