Download a copy of the vignette to follow along here: stability_measures.Rmd
In this vignette, we will highlight the main stability measure
options in the metasnf package.
Data set-up
library(metasnf)
my_dl <- data_list(
list(cort_t, "cortical_thickness", "neuroimaging", "continuous"),
list(cort_sa, "cortical_area", "neuroimaging", "continuous"),
list(subc_v, "subcortical_volume", "neuroimaging", "continuous"),
list(income, "household_income", "demographics", "continuous"),
list(pubertal, "pubertal_status", "demographics", "continuous"),
uid = "unique_id"
)
set.seed(42)
sc <- snf_config(
my_dl,
n_solutions = 4,
max_k = 40
)
sol_df <- batch_snf(my_dl, sc)To begin start calculating resampling-based stability measures, we’ll
build subsamples of the data list using the subsample_dl
function.
my_dl_subsamples <- subsample_dl(
my_dl,
n_subsamples = 50,
subsample_fraction = 0.85
)my_dl_subsamples contains a list of 50 subsamples of the
full data list. Each variation only has a random 85% of the original
observations. Once the subsamples of the data list have been created, a
cluster solution must be
batch_subsample_results <- batch_snf_subsamples(
my_dl_subsamples,
sc,
verbose = TRUE
)By default, the function returns a one-element list:
cluster_solutions, which is itself a list of cluster
solution data frames corresponding to each of the provided data list
subsamples. Setting the parameters return_sim_mats and
return_solutions to TRUE will turn the result
of the function to a three-element list containing the corresponding
solutions data frames and final fused similarity matrices of those
cluster solutions, should you require these objects for your own
stability calculations.
The function subsample_pairwise_aris can then be used to
calculate the ARIs between cluster solutions across the subsamples.
pairwise_aris <- subsample_pairwise_aris(
batch_subsample_results,
verbose = TRUE
)pairwise_aris is a list containing a summary data frame
of the ARIs between subsamples for each row of the original settings
data frame as well as another list of all the generated inter-subsample
ARIs as a result of setting return_raw_aris to
TRUE. The raw inter-subsample ARIs corresponding to a
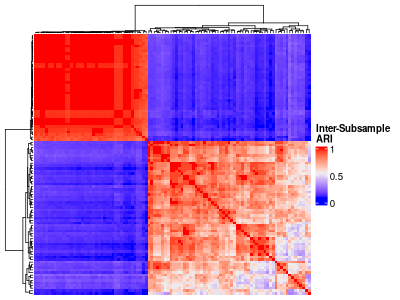
particualr settings data frame row can be visualized with a heatmap:
inter_ss_ari_hm <- ComplexHeatmap::Heatmap(
pairwise_aris$"raw_aris"$"s1",
heatmap_legend_param = list(
color_bar = "continuous",
title = "Inter-Subsample\nARI",
at = c(0, 0.5, 1)
),
show_column_names = FALSE,
show_row_names = FALSE
)
To calculate information about how often each pair of observations
clustered together across the subsamples, we can use the
calculate_coclustering function:
coclustering_results <- calculate_coclustering(
batch_subsample_results,
sol_df,
verbose = TRUE
)
coclustering_results$"cocluster_summary"The output of calculate_coclustering is a list
containing the following components: - cocluster_dfs: A
list of data frames, one per cluster solution, that shows the number of
times that every pair of observations in the original cluster solution
occurred in the same subsample, the number of times that every pair
clustered together in a subsample, and the corresponding fraction of
times that every pair clustered together in a subsample. -
cocluster_ss_mats: The number of times every pair of
observations occurred in the same subsample, formatted as a pairwise
matrix. - cocluster_sc_mats: The number of times every pair
of observations occurred in the same cluster, formatted as a pairwise
matrix. - cocluster_cf_mats: The fraction of times every
pair of observations occurred in the same cluster, formatted as a
pairwise matrix. - cocluster_summary: Among pairs of
observations that clustered together in the original cluster solution,
the mean fraction those pairs remained clustered together across the
subsample-derived solutions. This information is formatted as a data
frame with one row per cluster solution.
The cocluster_dfs component can be used to visualize
co-clustering across subsamples as a density plot:
cocluster_dfs <- coclustering_results$"cocluster_dfs"
cocluster_density(cocluster_dfs[[1]])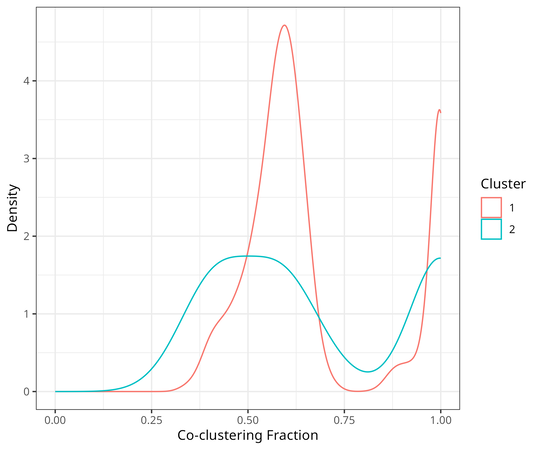
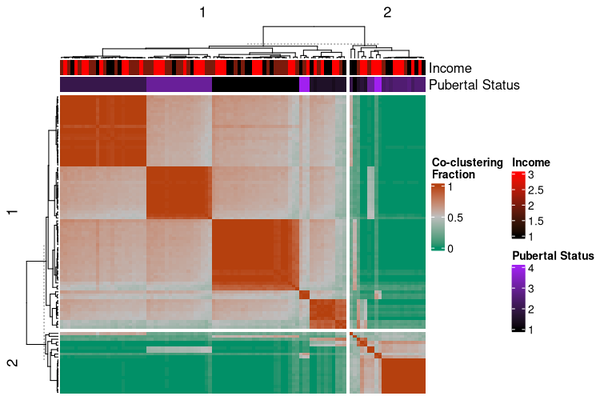
Or as a heatmap:
# Fraction of co-clustering between observations, grouped by original
# cluster membership
cocluster_heatmap(
cocluster_dfs[[1]],
dl = my_dl,
top_hm = list(
"Income" = "household_income",
"Pubertal Status" = "pubertal_status"
),
annotation_colours = list(
"Pubertal Status" = colour_scale(
c(1, 4),
min_colour = "black",
max_colour = "purple"
),
"Income" = colour_scale(
c(0, 4),
min_colour = "black",
max_colour = "red"
)
)
)